Research
Plant Immune System
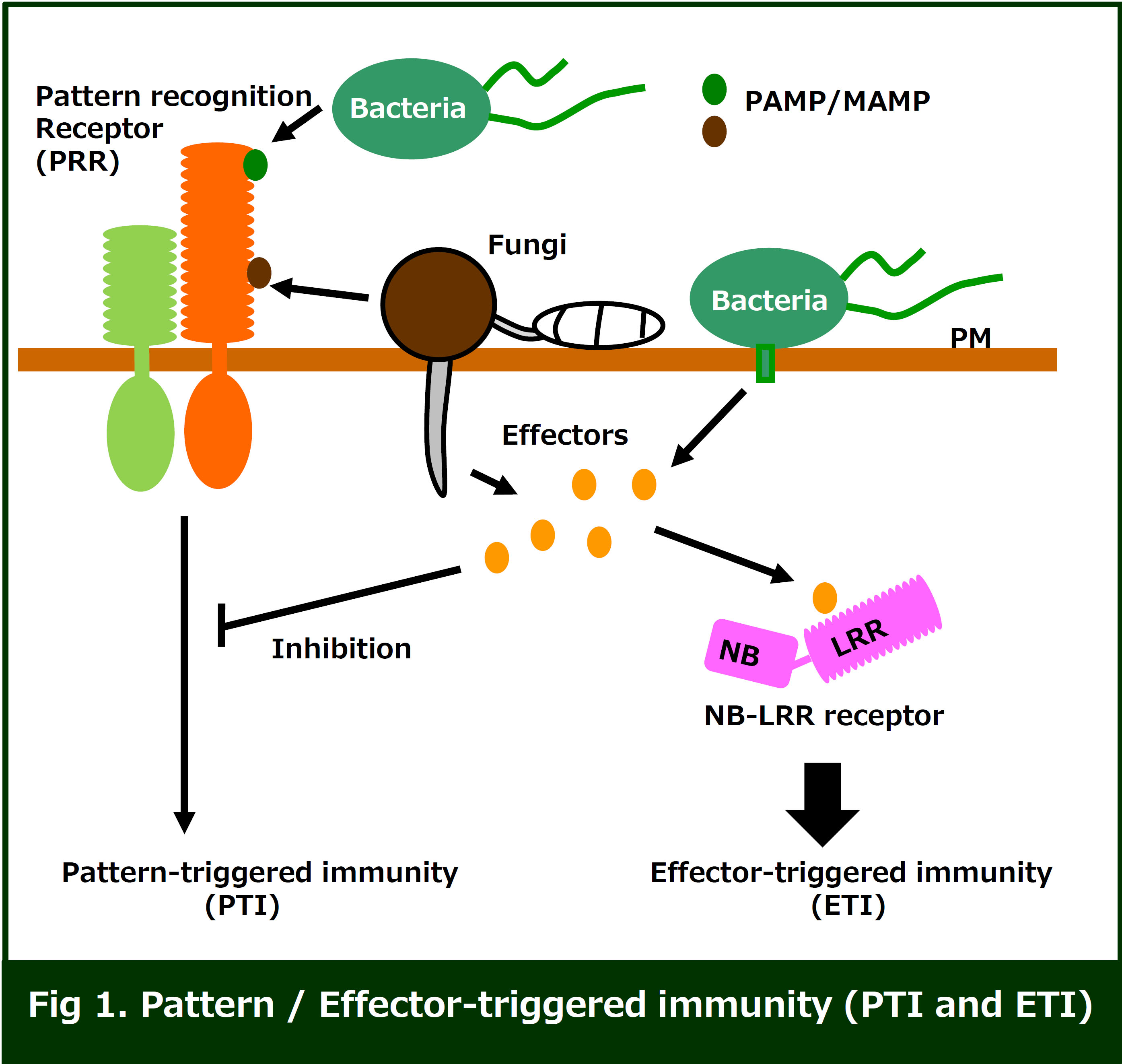
The plant innate immunity system is generally initiated via recognition
of conserved pathogen / microbe-associated molecular pattern (PAMP / MAMP)
ligands, including bacterial flagellin or peptidoglycan, and fungal chitin.
These MAMPs are perceived by pattern recognition receptors (PRRs), leading
to activation of a series of immune responses (PTI: pattern-triggered immunity).
PRRs are classified into two types of receptors, receptor-like kinase (RLKs)
and receptor-like proteins (RLP). Both types of PRRs contain ligand-binding
domains and a transmembrane domain, but only RLKs have an intercellular
kinase domain. The well-characterized ligand-binding domains are LRR (leucine-rich
repeat) and LysM (lysin rich motif).
Successful pathogens are able to secrete or directly deliver effector
proteins into the host cytoplasm which results in inhibition of host PTI.
To overcome PTI inhibition by the effectors, plants have developed intracellular
immune receptors of the nucleotide-binding leucine-rich repeat (NB-LRR)
protein family, which directly or indirectly recognize the effectors, and
activate strong immune responses (ETI: effector-triggered immunity).
Perception of PAMPs with PRRs induces a rapid intracellular activation
of mitogen-activated protein kinase (MAPK) cascades consisting of highly
conserved modules in eukaryotes. Each MAPK cascade is composed of three
consecutively acting protein kinases: a MAPK kinase kinase (MAPKKK), a
MAPK kinase (MAPKK) and a MAPK. The MAPK cascades induce robust immune
responses by phosphorylation of multiple transcription factors.
Current Projects
1)Elucidation of molecular mechanism of pattern-triggered immunity in plants.
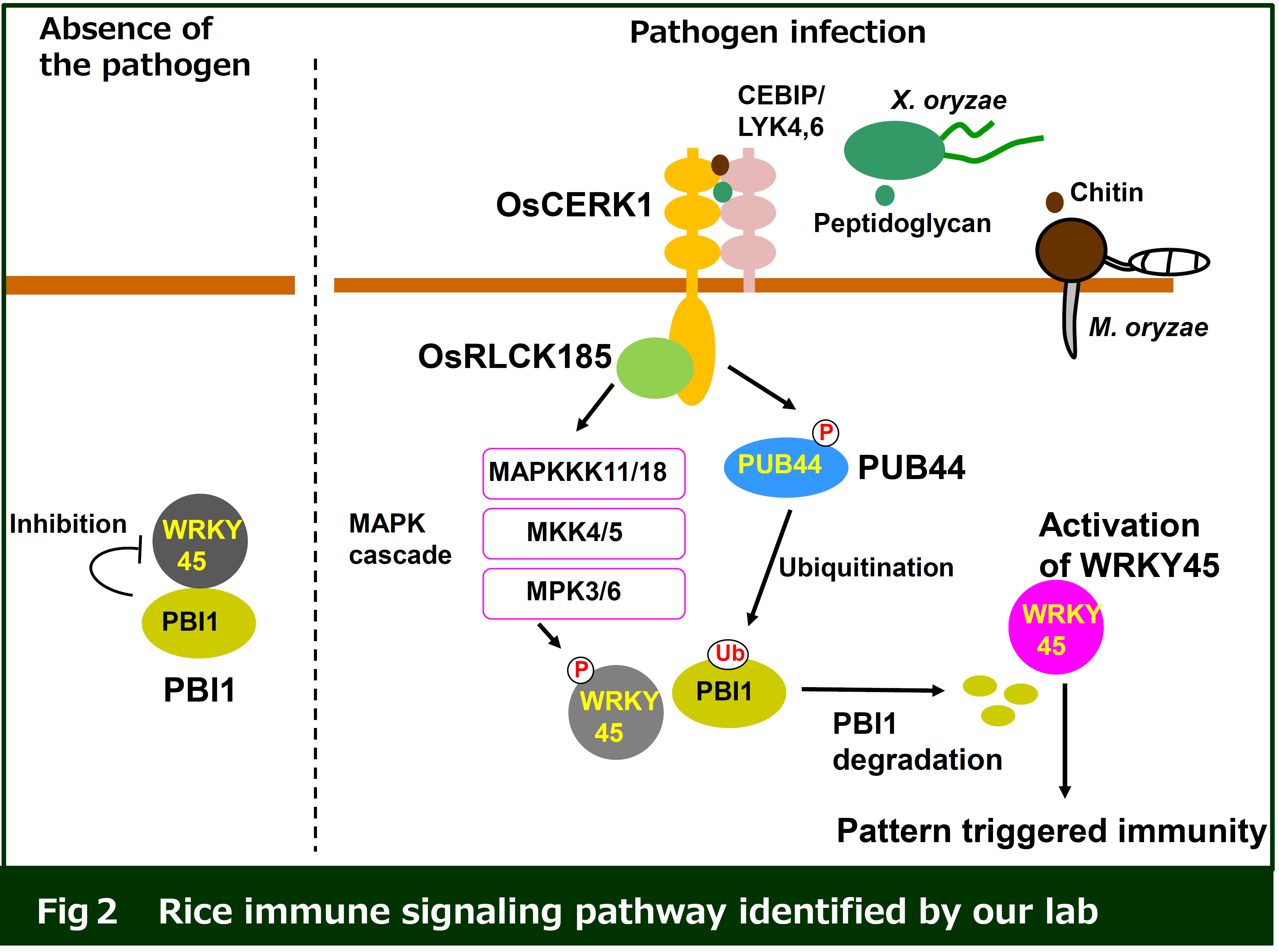
PRRs recognize PAMPs at the plasma membrane, which triggers an intracellular immune response. However, the molecular mechanisms remain to be identified. Our research focuses on immune signaling induced by the recognition of fungal chitin and bacterial peptidoglycan. Rice CEBiP and LYP4/LYP6 are RLPs with extracellular LysM domains responsible for recognition of chitin and PGN, respectively. Perception of chitin with CEBiP induces the formation of a complex with the RLK OsCERK1, which activates intracellular immune responses. We have identified that a receptor-like cytoplasmic kinase (RLCK), OsRLCK185, interacts with the cytoplasmic kinase domain of OsCERK1. In addition, chitin perception triggers OsCERK1-mediated phosphorylation of OsRLCK185, which activates a variety of immune responses, including MAP kinase activation (Cell Host Microbe 2013, Plant Cell Physiol 2017).
We identified PBL27, an Arabidopsis homolog of OsRLCK185. PBL27 interacts with CERK1 at plasma membrane and is phosphorylated by CERK1 in response to chitin. PBL27 also activates immune responses in a manner similar to OsRLCK185 (EMBO J 2016). There studies revealed that the RLCK family connects PRR-mediated perception to MAPK activation.
We found that the rice ubiquitin E3 ligase PUB44 plays an important role in pattern triggered immunity (Nature Communications 2014). In addition, we identified a novel immune factor, PBI1, as an interactor of PUB44. In the absence of pathogens, PBI1 interacts with and inhibits WRKY45, a key factor in rice immunity. PRR-mediated perception of PAMP/MAMP activates PUB44, which then ubiquitinates PBI1. The degradation of PBI1 results in the activation of WRKY45, which triggers various immune responses.
2)Understanding the inhibitory mechanisms by pathogen effectors
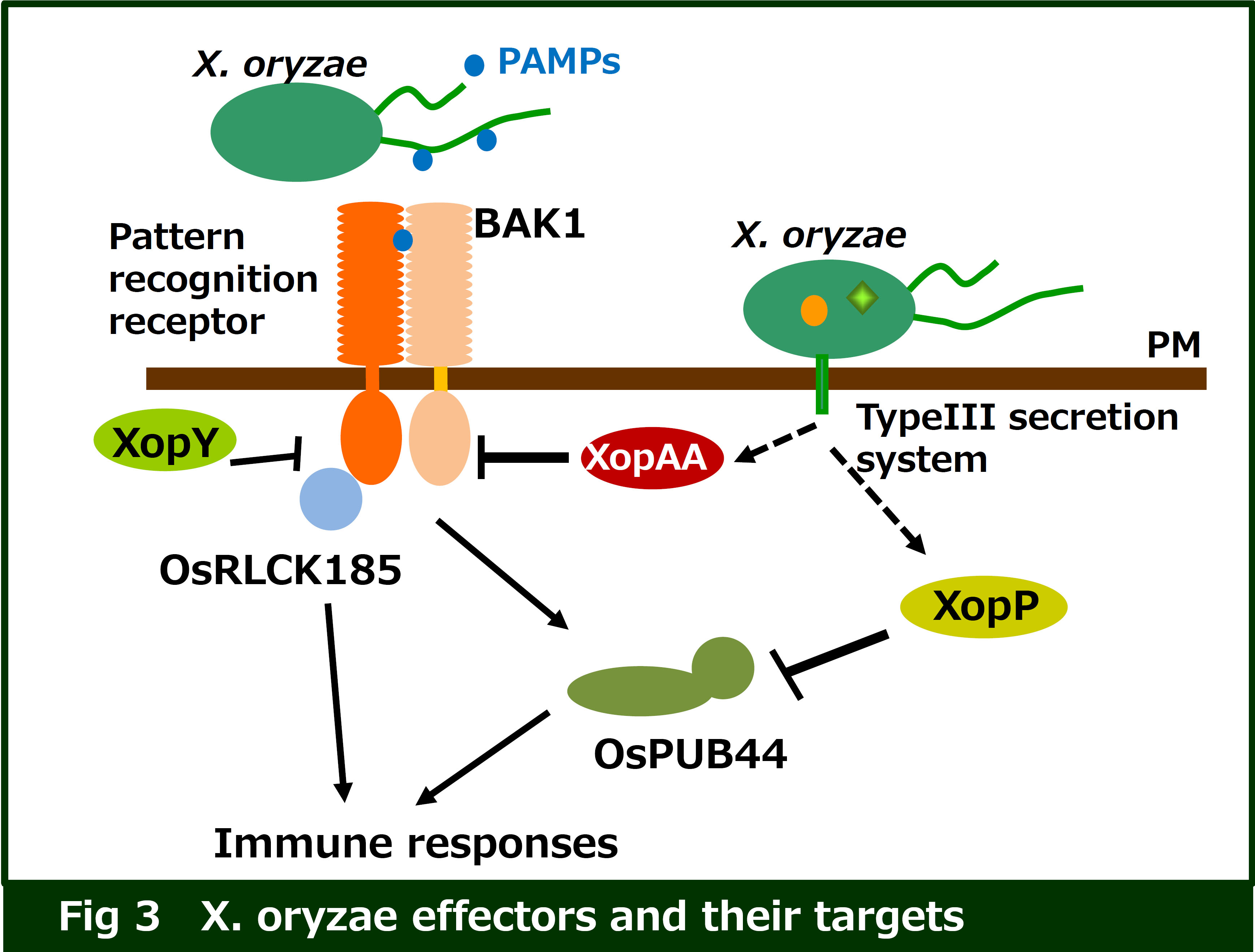
Pathogen effectors are secreted into host cells, and inhibit the immune responses. In fact, the expression of Xanthomonas oryzae pv. oryzae (Xoo) effectors in rice cells results in the inhibition of pattern-triggered immunity (PTI), suggesting that these effectors target critical immune factors. We have identified several immune factors interact with these effectors through yeast two-hybrid screening. We are currently analyzing the functions of these factors in the plant immune response, and the mechanisms by which pathogen effectors inhibit host immunity (Fig. 3)..
We have identified a rice receptor-like cytoplasmic kinase (OsRLCK185) that is targeted by Xoo effector XopY (Xoo1488). Our research indicates that XopY suppresses chitin-induced immunity by inhibiting the phosphorylation of OsRLCK185 by OsCERK1 (Cell Host Microbe 2013).
We found that another Xoo effector, XopP (Xoo3222) targets PUB44, a rice U-box type ubiquitin E3 ligases. PUB44 functions as a positive regulator in chitin- and peptidoglycan (PGN)-induced immunity. The U-box domain of PUB44 possesses the ubiquitin ligase activity. XopP interacts with the U-box domain, which results in suppression of the PUB44 enzymatic activity. These data indicate that XopP modulates host ubiquitination system to inhibit the immunity (Nature Communications 2014).
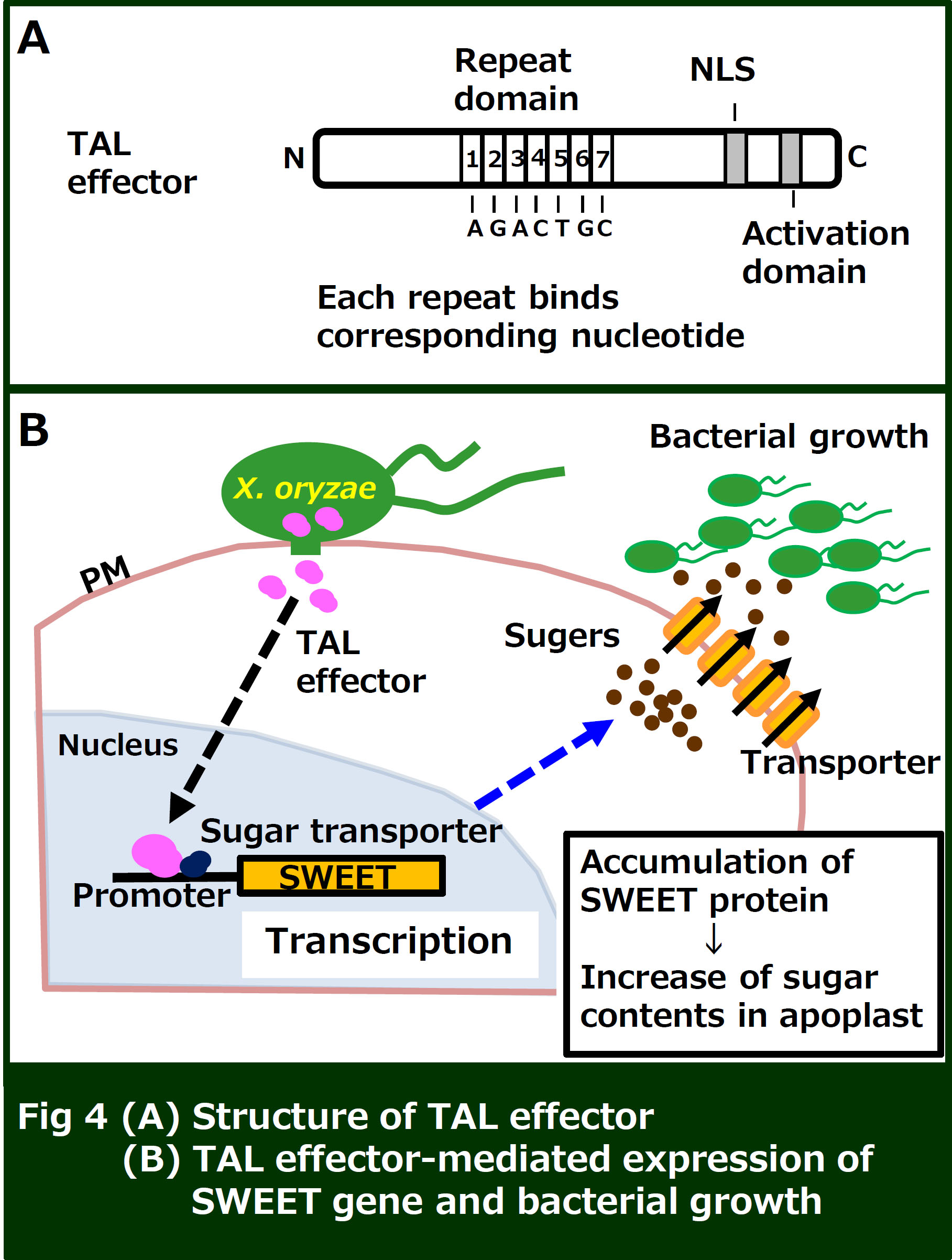
We are also analyzing Xoo TAL (Transcription Activator-Like) effector. TAL effector contains a unique DNA binding domain and functions as a transcription factor in rice nuclei. In rice, TAL effectors are known to activate the expression of sugar transporters (SWEET), which provide sugars for the pathogens residing in the apoplast. We are investigating how TAL effectors hijack the plant transcriptional system.
3)Elucidation of molecular mechanism of effector-triggered immunity in plants
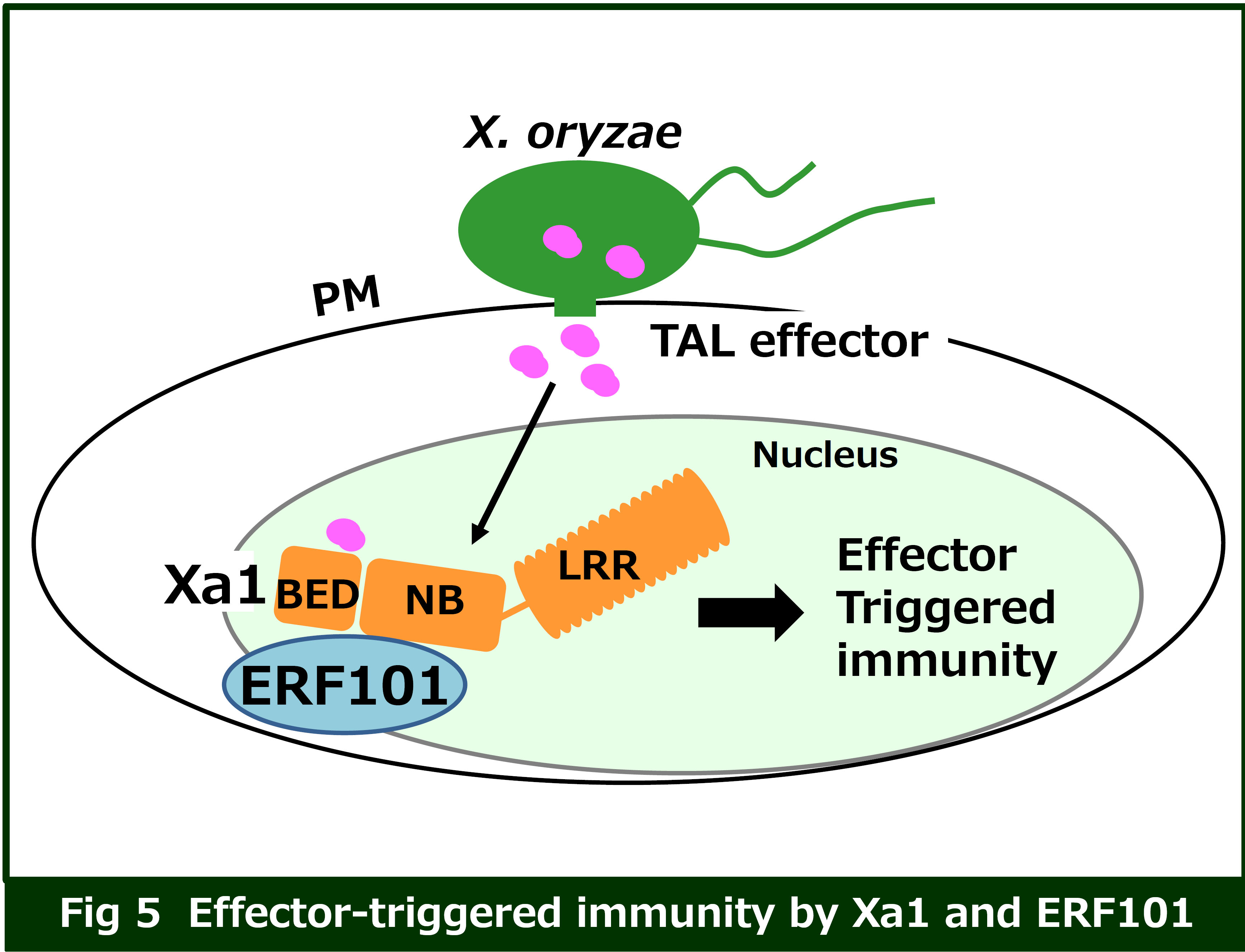
The intracellular NB-LRR receptors recognize effectors derived from bacterial and fungal pathogens, as well as insects, and induce a strong immune response. Recent research has discovered that NB-LRR receptors form higher-order protein complexes known as resistosome. However, the mechanism by which the NB-LRR receptors activate the immune response remains to be elucidated.
Rice Xa1 is one of the rice NB-LRR receptors that recognizes TAL effectors and induces immune responses. We have identified the AP2/ERF-type transcription factor ERF101 as an interactor of Xa1. Our findings suggest that ERF101 functions as a positive regulator in Xa1-mediated immunity. We are currently analyzing how the Xa1-ERF101 complex recognizes the TAL effector and activates effector-triggered immunity.
4)Understanding of immune signaling in plant
The perception of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) induces the activation of MAP kinases, the generation of reactive oxygen species (ROS), and calcium influx, which subsequently activate a variety of immune responses. So far, we have elucidated the molecular mechanisms by which OsRLCK185 and PBL27 activate the MAPK cascade downstream of PRRs (EMBO J 2016). We are currently investigating how rice factors targeted by Xoo effectors regulate immune signaling.
5) Research on symbiotic microorganisms controlling plant growth
Many microorganisms exist within plants and in the soil, supporting plant growth and immunity. Notable examples of symbiotic microorganisms include rhizobia and mycorrhizal fungi. In rice plants, the infection of mycorrhizal fungi is known to be regulated by OsCERK1. We are analyzing how the downstream components of OsCERK1 regulate these symbiotic responses. Additionally, we are studying newly identified symbiotic microorganisms that promote plant growth.
6) New technology to improve plant disease resistance
During the past several decades, plant Immune research has identified many important genes regulating host immunity from model plants such as Arabidopsis and rice. Recently, the genes were used to develop broad-spectrum and durable resistance to pathogens. This type of research is termed as ‘translational research.
